
1️⃣ Part 1: Supervised Learning & Neural Networks
2️⃣ Part 2: Randomized Optimization
3️⃣ Part 3: Unsupervised Learning & Dimensionality Reduction
4️⃣ (Coming soon)
Introduction
Unlike supervised learning—where models learn from labeled examples—unsupervised learning aims to uncover patterns in data without any ground-truth labels. The goal is to find natural groupings, meaningful structure, or lower-dimensional representations hidden inside high-dimensional datasets.
In this part of the Machine Learning series, I focused on two major unsupervised learning themes:
🧩 1. Clustering Algorithms
Clustering groups similar data points based on their position in feature space. The project evaluates two core methods:
- K-Means — partitions points into (k) compact clusters by minimizing distance to cluster centroids.
- Gaussian Mixture Models (GMM) — a probabilistic approach that models the data as a mixture of Gaussians, allowing soft membership and more flexible cluster shapes.
These methods help answer:
- Do natural subgroups exist in this dataset?
- How well separated—or overlapping—are the clusters?
🔻 2. Dimensionality Reduction
Dimensionality reduction (DR) transforms the data into a smaller set of informative features, often revealing structure that clustering alone can’t capture. The techniques explored include:
- PCA — captures directions of highest variance
- ICA — separates independent latent signals
- Random Projection — compresses features while preserving distances
- Random Forest feature selection — keeps only the most informative features
These DR methods are then combined with clustering—and later, a neural network—to evaluate how reduced representations affect performance.
Two datasets were used:
- 🍷 Wine Quality — chemical properties predicting quality
- 🐚 Abalone — physical measurements predicting age
Part 1: Baseline Clustering Performance
Before applying dimensionality reduction, I evaluated how K-Means and GMM perform on the raw datasets.
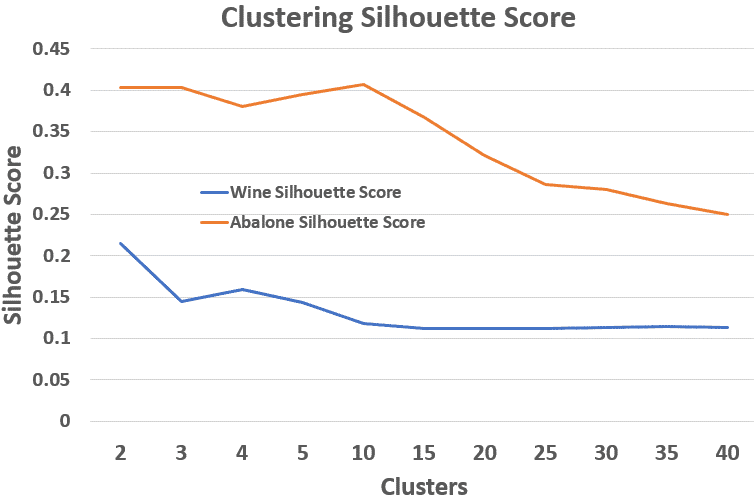
A consistent pattern emerged: As the number of clusters increases, the silhouette score — which measures how well a point fits within its own cluster compared to the nearest other cluster — tends to drop, since clusters naturally become more compressed.
Abalone – Baseline Silhouette Scores
Part 2: Dimensionality Reduction
Each dimensionality reduction method reshapes the feature space differently:
- PCA rotates the data to maximize variance retention
- ICA attempts to separate independent signals
- Random Projection (RP) compresses dimensionality with JL guarantees
- Random Forest (RF) selects features based on importance (Gini)
The goal was to see whether these transforms help clustering uncover more meaningful patterns, or reduce noise that might blur cluster boundaries.
📉 t-SNE Visualization (Exploratory)
Although not used directly for clustering or neural networks, t-SNE (t-Distributed Stochastic Neighbor Embedding) is a nonlinear technique for visualizing high-dimensional structure in 2D. It preserves local relationships, making it useful for checking whether a dataset has visually separable clusters before performing formal analysis.
Example – Abalone t-SNE 2D Plot

t-SNE reveals why Abalone is difficult to cluster: the classes heavily overlap, even under a nonlinear embedding. This visual intuition aligns with the modest improvements seen from PCA, RP, and RF later in the analysis.
Part 3: Clustering on Reduced Dimensions
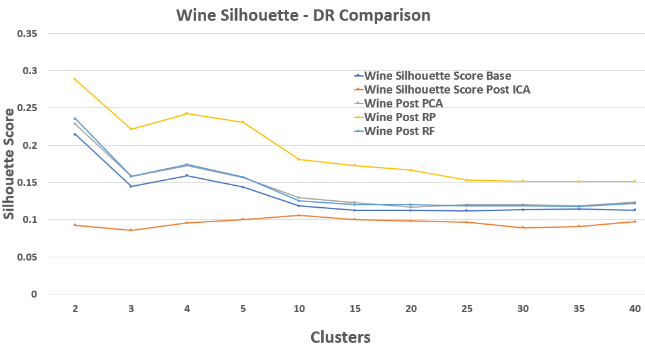
Wine – Silhouette Score Comparison
Random Projection surprisingly produces the most distinct clusters early on, likely because it breaks noisy feature correlations.
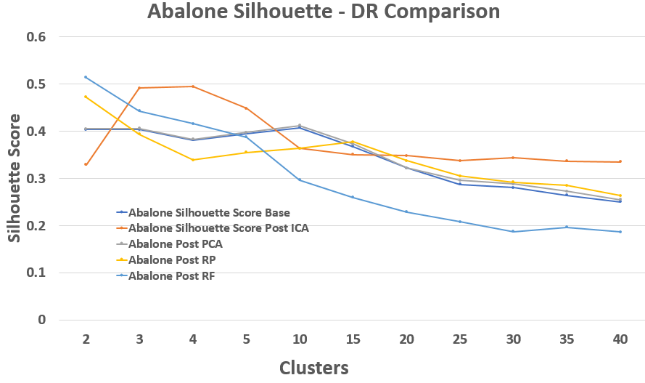
Abalone – Silhouette Score Comparison
Abalone is noisier and lower-dimensional, so improvement is more modest, but ICA consistently underperforms.
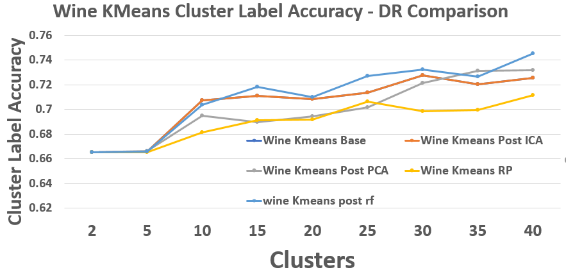
Wine – K-Means Cluster Label Accuracy
Random Forest feature selection slightly outperforms other DR methods, suggesting that filtering noisy features helps more than rotating or projecting them.
Part 4: GMM Performance on Reduced Data
Gaussian Mixture Models capture cluster shape better than K-Means, especially where clusters overlap — like in the Wine dataset.
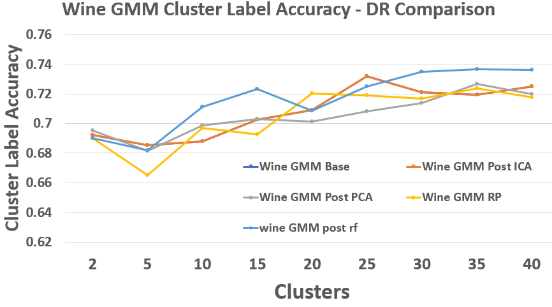
Wine – GMM Label Accuracy
RF again yields the strongest or near-strongest accuracy across cluster sizes.
Part 5: Abalone – DR Performance Highlights
While Wine benefits noticeably from DR, Abalone is lower-dimensional and noisier, meaning differences across DR methods were smaller.
The one clear takeaway: ICA consistently performs the worst, both in silhouette and label accuracy.
Abalone – Silhouette Comparison With DR
Visible struggle from ICA compared to PCA, RP, or RF.

Part 6: Neural Network Performance on DR + Cluster Features
I trained a small neural network using:
- Original features
- PCA-reduced features
- ICA-reduced features
- RP-reduced features
- RF-selected features
- Cluster assignments as features (K-Means labels)
Two key observations:
-
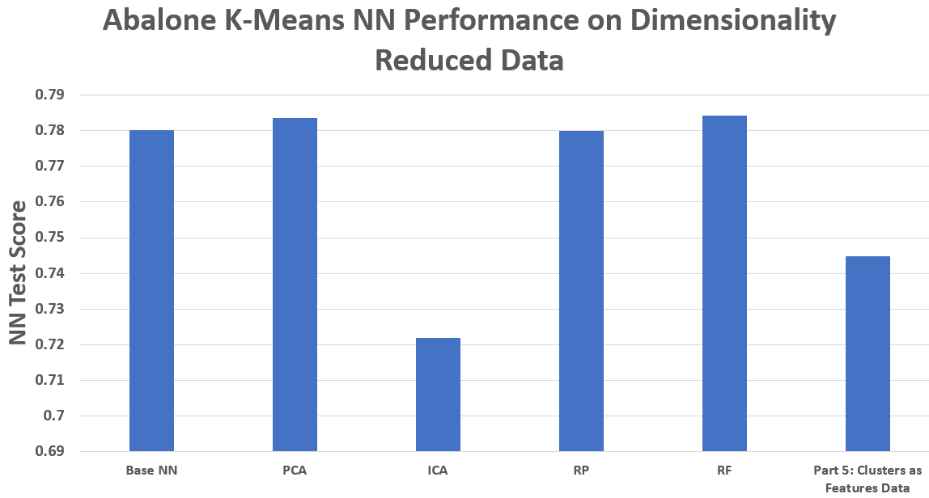
RF-selected features almost always gave the best NN test accuracy. Removing noisy or irrelevant inputs proved more effective than transforming all features.
-
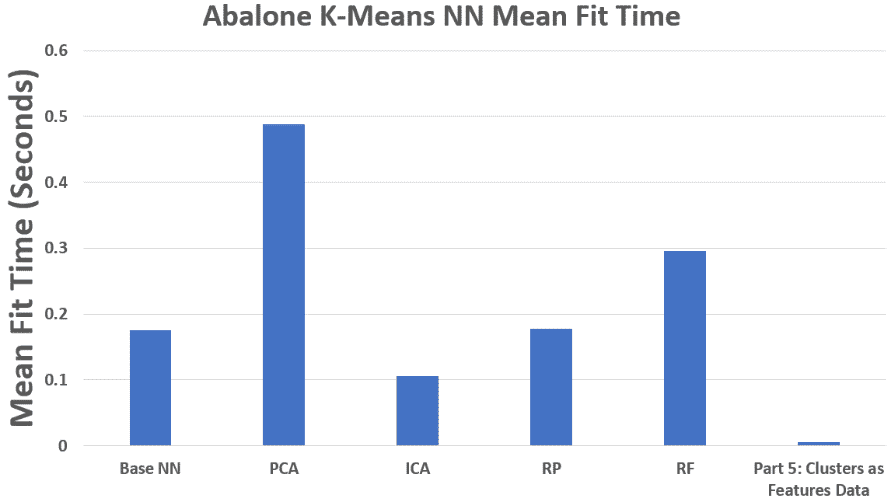
Cluster labels as features yielded the fastest runtime but weaker accuracy, since cluster IDs alone discard too much information.
Abalone – NN Performance on DR Feature Sets
Abalone – NN Mean Fit Time
Conclusion
Across both datasets, dimensionality reduction had mixed but insightful effects:
🔹 Wine Dataset 🍷
- RF feature selection provided the strongest results for both K-Means and GMM.
- Random Projection produced the highest silhouette scores early on.
- Dimensionality reduction clearly improved cluster stability and separability.
🔹 Abalone Dataset 🐚
- Much less improvement across techniques — the dataset is noisy and nearly low-dimensional already.
- ICA performed the worst across all metrics.
- RF and PCA showed small but consistent gains.
🔹 Neural Networks on Reduced Data
- RF-selected features delivered the best test accuracy.
- Cluster labels were extremely fast but cost accuracy.
🧠 Final Takeaway
This part of the project reinforced the No Free Lunch Theorem: No single dimensionality reduction method is universally best.
Performance depends heavily on dataset structure:
- 🍷 Wine’s correlated chemical signals → DR reveals clearer structure
- 🐚 Abalone’s noisy, compact feature space → limited benefit from DR